Note:
GPRsurvey
However, this tutorial will not explain you the math/algorithms behind the different processing methods.
We will work with the data contained in the directory
/exampleDataCube/Grid-dir1-Rawdata/.
if(!require("devtools")) install.packages("devtools")
devtools::install_github("emanuelhuber/RGPR")
library(RGPR) # load RGPR in the current R session
[optionally] If RGPR is not installed, follow the instructions of
the tutorial “Getting started” to install it.
The working directory must be correctly set to use relative filepaths. The working directory can be set either in your R-software or in R directly with (of course you need to adapt the filepath shown below to your system):
DIR <- file.path("your_dir_path/GPRdata-master/exampleDataCube")
setwd(DIR) # set the working directory
getwd() # Return the current working directory (just to check)
GPRsurveyAn object of the class GPRsurvey is like an index that contains some
of the meta-data of several GPR data recorded during one survey. With
the class GPRsurvey you have an overview of all your data, you can
compute the positions of the profile intersections, plot a top view of
the survey and plot the data in 3D with open-GL (implemented in the
R-package RGL).
Read all the GPR records (“.DZT”) located in the directory
/exampleDataCube/Grid-dir1-Rawdata/
LINES <- file.path( paste0("FILE____", sprintf("%03d", 1:46), ".DZT"))
sprintf format the integer values ranging from 1 to 46 such that they
match the file names (e.g., 1 becomes "001", 2 becomes "002", …, 46
becomes "046"). The 0 in "%03d" means pad with zeros to the field,
3 means that the field length is 3, d that the values are integer.
GPRsurveySU <- GPRsurvey(LINES, verbose = FALSE)
Have a look at the newly created object:
SU
## *** Class GPRsurvey ***
## Unique directory:.
## - - - - - - - - - - - - - - -
## name length units date freq coord int filename
## 1 FILE____001 9.0 m 2013-10-29 400 NO NO FILE____001.DZT
## 2 FILE____002 9.0 m 2013-10-29 400 NO NO FILE____002.DZT
## 3 FILE____003 9.0 m 2013-10-29 400 NO NO FILE____003.DZT
## 4 FILE____004 9.0 m 2013-10-29 400 NO NO FILE____004.DZT
## 5 FILE____005 9.0 m 2013-10-29 400 NO NO FILE____005.DZT
## 6 FILE____006 9.0 m 2013-10-29 400 NO NO FILE____006.DZT
## 7 FILE____007 9.0 m 2013-10-29 400 NO NO FILE____007.DZT
## 8 FILE____008 9.0 m 2013-10-29 400 NO NO FILE____008.DZT
## 9 FILE____009 9.0 m 2013-10-29 400 NO NO FILE____009.DZT
## 10 FILE____010 9.0 m 2013-10-29 400 NO NO FILE____010.DZT
## 11 FILE____011 9.0 m 2013-10-29 400 NO NO FILE____011.DZT
## 12 FILE____012 9.0 m 2013-10-29 400 NO NO FILE____012.DZT
## 13 FILE____013 9.0 m 2013-10-29 400 NO NO FILE____013.DZT
## 14 FILE____014 9.0 m 2013-10-29 400 NO NO FILE____014.DZT
## 15 FILE____015 9.0 m 2013-10-29 400 NO NO FILE____015.DZT
## 16 FILE____016 9.0 m 2013-10-29 400 NO NO FILE____016.DZT
## 17 FILE____017 9.0 m 2013-10-29 400 NO NO FILE____017.DZT
## 18 FILE____018 9.0 m 2013-10-29 400 NO NO FILE____018.DZT
## 19 FILE____019 9.0 m 2013-10-29 400 NO NO FILE____019.DZT
## 20 FILE____020 9.0 m 2013-10-29 400 NO NO FILE____020.DZT
## 21 FILE____021 9.0 m 2013-10-29 400 NO NO FILE____021.DZT
## 22 FILE____022 9.0 m 2013-10-29 400 NO NO FILE____022.DZT
## 23 FILE____023 9.0 m 2013-10-29 400 NO NO FILE____023.DZT
## 24 FILE____024 9.0 m 2013-10-29 400 NO NO FILE____024.DZT
## 25 FILE____025 9.0 m 2013-10-29 400 NO NO FILE____025.DZT
## 26 FILE____026 9.0 m 2013-10-29 400 NO NO FILE____026.DZT
## 27 FILE____027 9.0 m 2013-10-29 400 NO NO FILE____027.DZT
## 28 FILE____028 9.0 m 2013-10-29 400 NO NO FILE____028.DZT
## 29 FILE____029 9.0 m 2013-10-29 400 NO NO FILE____029.DZT
## 30 FILE____030 9.0 m 2013-10-29 400 NO NO FILE____030.DZT
## 31 FILE____031 9.0 m 2013-10-29 400 NO NO FILE____031.DZT
## 32 FILE____032 9.0 m 2013-10-29 400 NO NO FILE____032.DZT
## 33 FILE____033 9.0 m 2013-10-29 400 NO NO FILE____033.DZT
## 34 FILE____034 9.0 m 2013-10-29 400 NO NO FILE____034.DZT
## 35 FILE____035 9.0 m 2013-10-29 400 NO NO FILE____035.DZT
## 36 FILE____036 9.0 m 2013-10-29 400 NO NO FILE____036.DZT
## 37 FILE____037 9.0 m 2013-10-29 400 NO NO FILE____037.DZT
## 38 FILE____038 9.0 m 2013-10-29 400 NO NO FILE____038.DZT
## 39 FILE____039 9.0 m 2013-10-29 400 NO NO FILE____039.DZT
## 40 FILE____040 9.0 m 2013-10-29 400 NO NO FILE____040.DZT
## 41 FILE____041 9.0 m 2013-10-29 400 NO NO FILE____041.DZT
## 42 FILE____042 9.0 m 2013-10-29 400 NO NO FILE____042.DZT
## 43 FILE____043 9.0 m 2013-10-29 400 NO NO FILE____043.DZT
## 44 FILE____044 9.0 m 2013-10-29 400 NO NO FILE____044.DZT
## 45 FILE____045 9.0 m 2013-10-29 400 NO NO FILE____045.DZT
## 46 FILE____046 9.0 m 2013-10-29 400 NO NO FILE____046.DZT
## ****************
You can see that no coordinates (x,y,z) are associated with the GPR data. Therefore, if you try to plot the survey you will get:
plot(SU, asp = 1) # throw an error
The data come without coordinates. To add coordinate to GPR data, check the tutorial Adding coordinates to GPR data. Note that RPGR automatically reads GPS files associated with the GPR data and interpolate the trace positions.
Here, we assume that the data were collected on a grid: all the GPR data are parallel, oriented in the same direction and spaced by 0.20 m. For grid settings, a new approach is here introduced (here only with data recorded along the y-direction, but it works also for data recorded along the x-direction and along both direction). If your data already have coordinates, skip the section below.
For this approach, all the data must be oriented in the same direction
(the x-lines must have the same direction, the y-lines must have the
same direction). If this is not the case for your data, you can use the
function reverse() to reverse the GPR line. You can specify which
lines must be reversed:
SU <- reverse(SU, id = seq(from = 2, to = 11, by = 2))
or if your data were collected in zig-zag (two adjacent GPR lines have
opposite direction), you can use the argument "zigzag":
#all the even GPR lines are reversed
SU <- reverse(SU, id = "zigzag")
To set the grid coordinates, use the function setGridCoord() and
assign the grid specifications in the form of a list. This list takes
for arguments (see also:
xlines: integer values corresponding to the x-lines in the
GPRsurvey object (if there are no x-lines, no need to specify
xlines).xpos: the position of the x-lines along the x-axis, same length as
xlines (if there are no x-lines, no need to specify xpos).xstart[optional]: shift to apply along the y-position, useful
if the lines do not start at the same position; same length as
xlines (if there are no x-lines, no need to specify xstart).xlength[optional]: the length of the lines. Note that normally
RGPR reads the line length from the data (if there are no x-lines,
no need to specify xlength).ylines: integer values corresponding to the x-lines in the
GPRsurvey object (if there are no y-lines, no need to specify
ylines).ypos: the position of the x-lines along the x-axis, same length as
ylines (if there are no y-lines, no need to specify ypos).ystart[optional]: shift to apply along the x-position, useful
if the lines do not start at the same position; same length as
ylines (if there are no y-lines, no need to specify ystart).ylength[optional]: the length of the lines. Note that normally
RGPR reads the line length from the data (if there are no y-lines,
no need to specify ylength).Note that:
xlines, xpos, xstart and xlength must be equal
(except if you omit xstart and/or xlength)ylines, ypos, ystart and ylength must be equal
(except if you omit ystart and/or ylength)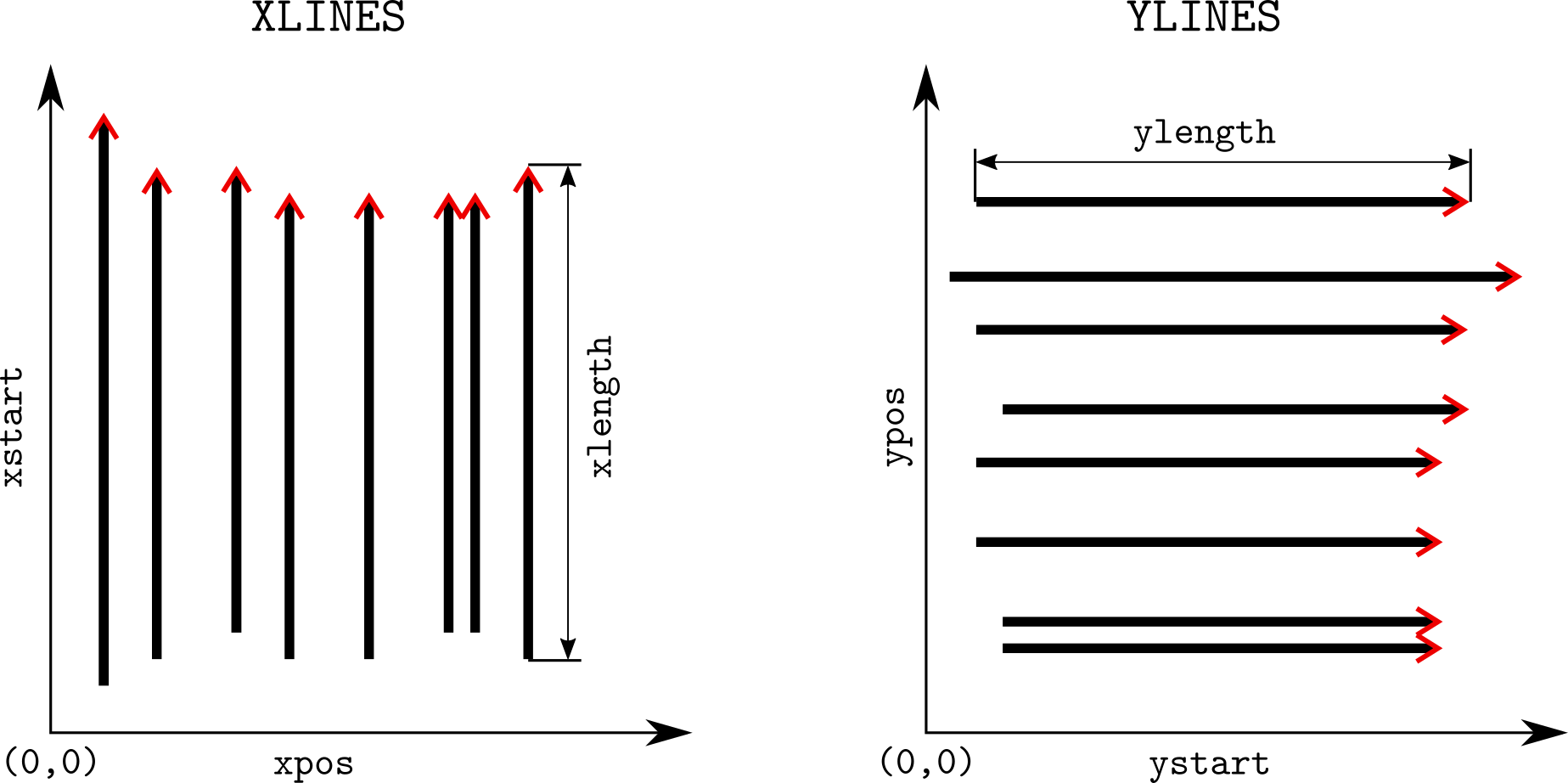
Imagine your data were collected as follows:
SU_img <- SU
xstart <- rep(0, 40)
xstart[c(3, 5)] <- 1
setGridCoord(SU_img) <- list(xlines = 1:40,
xpos = seq(0, by = 2, length.out = 40),
xstart = xstart,
ylines = 40 + (1:6),
ypos = c(0, 1, 2, 4, 6, 7.6),
ystart = c(-2, 0, 0, 0, 0, 0))
plot(SU_img, asp = TRUE, parFid = NULL)

Imagine your data were collected on a 19 m x 25m grid:
SU_img <- SU
setGridCoord(SU_img) <- list(xlines = 1:20,
xpos = seq(0, by = 1, length.out = 20),
xstart = rep(0, 20), # could be omitted
xlength = rep(25, 20),
ylines = 21:46,
ypos = seq(0, by = 1, length.out = 26),
ystart = rep(0, 26), # could be omitted
ylength = rep(19, 26))
plot(SU_img, asp = TRUE, parFid = NULL)

In our case we have only x-lines, so we don’t specify ylines, ypos,
and ystart. The 5th, 8th, 22th lines starts 1 m after the others; the
1st, 28th, 33rd, 38th, and 44th lines start 0.4 m after the others:
# define xstart
xstart <- rep(0, length(SU))
xstart[c(5, 8, 22)] <- 1
xstart[c(1, 28, 33, 38, 44)] <- 0.4
setGridCoord(SU) <- list(xlines = seq_along(SU),
xpos = seq(0, by = 0.2,
length.out = length(SU)),
xstart = xstart)
Now you can plot your survey data (because you have coordinates)
plot(SU, asp = TRUE, parFid = NULL)

We set parFid = NULL because we do not want to plot all the fiducial
markers
Plot a single GPR data with
plot(SU[[1]])
## Antenna separation set to 0 m. Set it with 'antsep(x) <-... '

You see that some processing is needed.
We apply some basic processing steps sequentially (estimate time-zero
and shift all the traces to time-zero, dewow, AGC-gain). Maybe you want
to horizontally smooth the data (un-comment the line starting
traceStat) or to interpolate the signal envelope (un-comment the line
starting envelope). For more info on GPR data processing check the
tutorial Basic GPR data
processing
SU <- papply(SU,
prc = list(estimateTime0 = list(method = "coppens", w = 2),
# "NULL" because we take the default
time0Cor = NULL,
dewow = list(w = 3),
gain = list(type = "agc", w = 1.2) #,
# traceStat = list(w = 20, FUN = mean),
# envelope = NULL)
))
Does it look better now?
plot(SU[[1]])

Now that the data are well prepared, the interpolation is a simple task.
We define the grid resolution in all three direction: dx = 0.05 m, dy =
0.05 m, dz = 0.05 ns as well as an additional parameter h (>0) that
controls the smoothness of the interpolation (the interpolation used is
the Multilevel B-spline Approximation as implemented in the function
mba.surf() of the package MBA).
SXY <- interpSlices(SU, dx = 0.05, dy = 0.05, dz = 0.05, h = 6)
SXY
## *** Class GPRcube ***
## dim: 199 x 219 x 901
## res: 0.05 m x 0.05 m x 0.05 ns
## extent: 9.9 m x 10.9 m x 45 ns
## crs:
## *********************
GPRcube objectSXY can be manipulated like an array with x, y and z dimensions.
Plot an horizontal slice of the data cube:
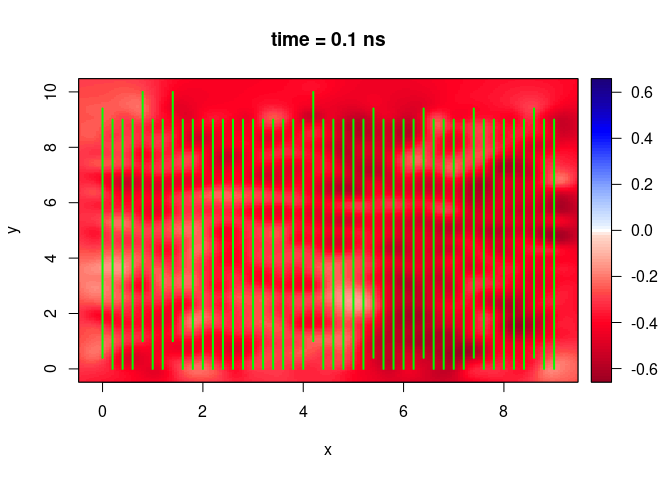
plot(SXY[,,10])

Plot a vertical slice along x-direction
plot(SXY[,10,])

Plot a vertical slice along y-direction
plot(SXY[10,,])

You can define the same color range for each plot:
# color range (over all possible slice values)
clim <- range(SXY, na.rm = TRUE)
plot(SXY[,,50], clim = clim)

You can change the color palette? displayPalGPR() shows all the
palette available in RGPR
displayPalGPR()

Try these two palettes: "sunny" and "slice":
plot(SXY[,,50], clim = clim, col = palGPR("sunny"), asp = 1)

plot(SXY[,,50], clim = clim, col = palGPR("slice"), asp = 1)

By defaut the slices are interpolated whithin the convex hull of the GPR lines that is extended by 5 percents.
It is possible to specify the type of interpolation extend with the
argument extend that can take the following values:
chull: the convex-hullbbox: the axis-aligned bounding boxobbox: the oriented bounding boxbuffer: an area around the GPR lines (like a buffer). In this case
a buffer value> 0 must be defined.Below are some examples (the case extend = "obbox" makes here no sense
because the oriented bounding-box is the same as the axis-aligned
bounding box).
SXY <- interpSlices(SU, dx = 0.05, dy = 0.05, dz = 0.05, h = 6, extend = "chull", buffer = 0)
plot(SXY[,,10])
lines(SU, col = "green", lwd = 2)
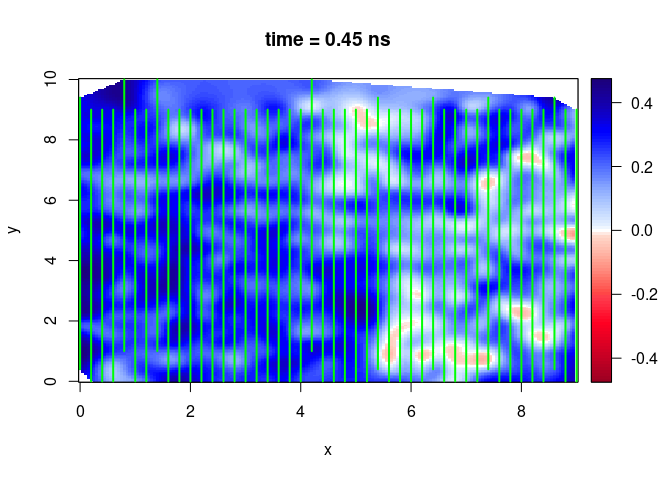
SXY <- interpSlices(SU, dx = 0.05, dy = 0.05, dz = 0.05, h = 6, extend = "chull", buffer = 2)
plot(SXY[,,10])
lines(SU, col = "green", lwd = 2)
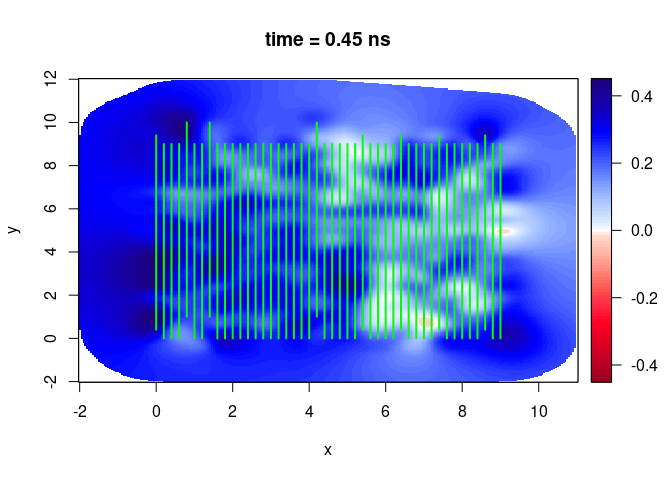
SXY <- interpSlices(SU, dx = 0.05, dy = 0.05, dz = 0.05, h = 6, extend = "bbox")
plot(SXY[,,3])
lines(SU, col = "green", lwd = 2)
SXY <- interpSlices(SU, dx = 0.05, dy = 0.05, dz = 0.05, h = 6, extend = "bbox", buffer = 3)
plot(SXY[,,10])
lines(SU, col = "green", lwd = 2)
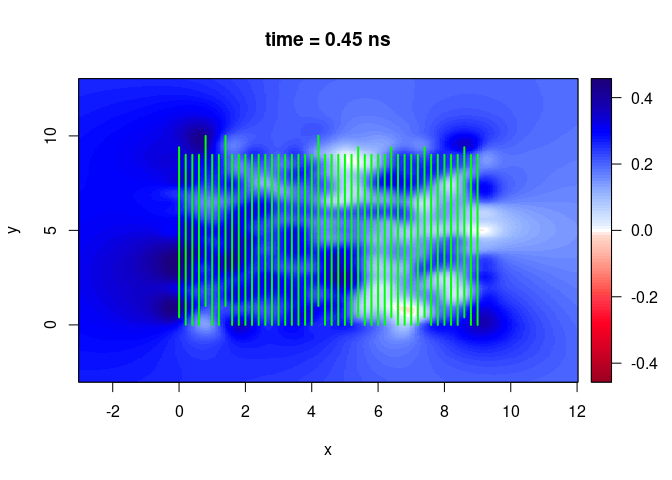
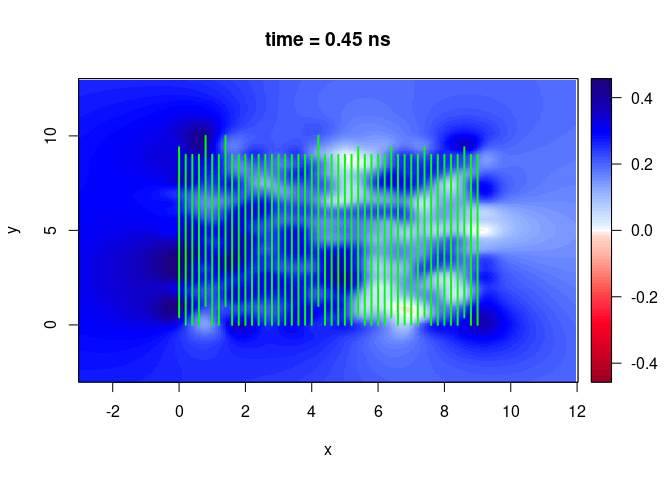
SXY <- interpSlices(SU, dx = 0.05, dy = 0.05, dz = 0.05, h = 6, extend = "obbox")
plot(SXY[,,10])
lines(SU, col = "green", lwd = 2)
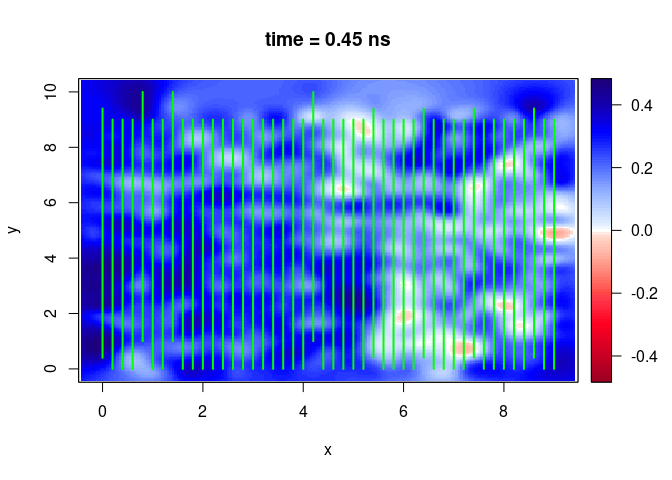
SXY <- interpSlices(SU, dx = 0.05, dy = 0.05, dz = 0.05, h = 6, extend = "obbox", buffer = 3)
plot(SXY[,,10])
lines(SU, col = "green", lwd = 2)
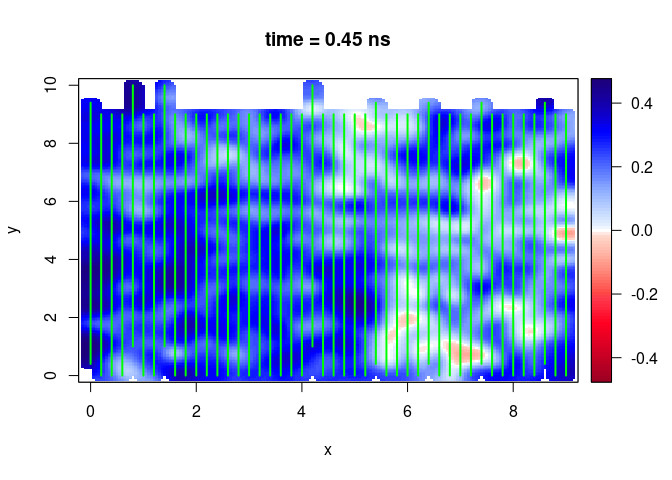
SXY <- interpSlices(SU, dx = 0.05, dy = 0.05, dz = 0.05, h = 6, extend = "buffer", buffer = 0.2)
plot(SXY[,,10])
lines(SU, col = "green", lwd = 2)
You first need to convert the slice you want to export into a raster
object in R (defined in the raster package):
r <- as.raster(SXY[,,10])
## The legacy packages maptools, rgdal, and rgeos, underpinning the sp package,
## which was just loaded, will retire in October 2023.
## Please refer to R-spatial evolution reports for details, especially
## https://r-spatial.org/r/2023/05/15/evolution4.html.
## It may be desirable to make the sf package available;
## package maintainers should consider adding sf to Suggests:.
## The sp package is now running under evolution status 2
## (status 2 uses the sf package in place of rgdal)
Then, use the writeRaster() function of the raster package to export
the slice in the raster format you like (check the help on this
function, ?raster::writeRaster):
raster::writeRaster(r, filename = "slice10.tif")
You may also want to export the coordinates as shapefiles or geodata, as lines
exportCoord(SU, type = c("SpatialLines"), fPath = "myshapefile.shp",
driver = "ESRI Shapefile")
… or as points
exportCoord(SU, type = c("SpatialPoints"), fPath = "myshapefile.shp",
driver = "ESRI Shapefile")
Adapt the driver and the filename extension to your need (for that,
check the help on the rgdal::writeOGR() function).